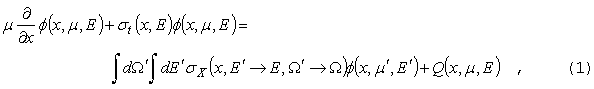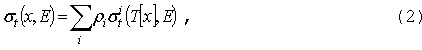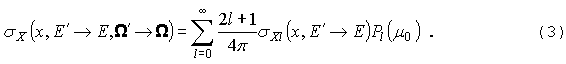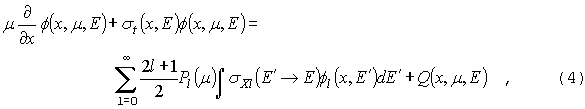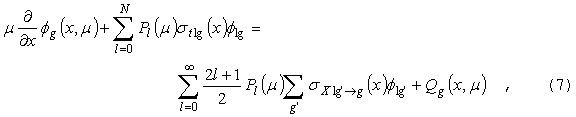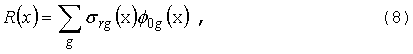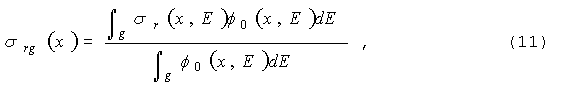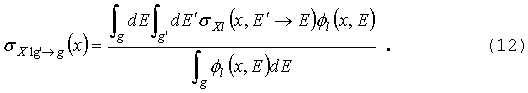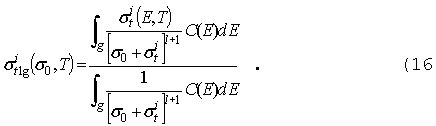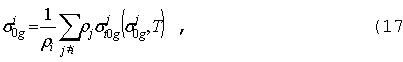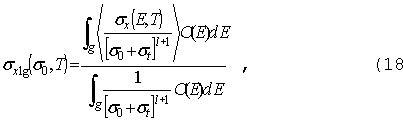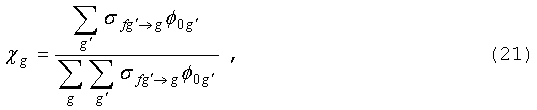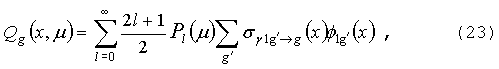GNPDL-30/19:
БИБЛИОТЕКА НЕЙТРОННО-ФОТОННЫХ ГРУППОВЫХ КОНСТАНТ
ДЛЯ РАСЧЕТА ПОДКРИТИЧЕСКИХ СИСТЕМ С ВНЕШНЕЙ ПОДСВЕТКОЙ
|
|
|
где
f
- поток, зависящий от координаты x, m
- косинус полярного угла для направления W,
Е - энергия ( для упрощения формул
рассмотрение ведется в одномерной плоской геометрии). Полное сечение st
предполагается зависящим от координаты и энергии. Правая часть уравнения
описывает источник, состоящий из двух компонент: внутренний, обусловленный
образованием нейтронов с направлением W
и энергией E в результате различных
взаимодействий с ядрами нейтронов с направлением движения Wў
и энергией Eў (вероятность
этого процесса характеризуется "сечением переноса" sX )и
внешний, независимый источник Q.
|
|
Макроскопические сечения (в единицах
см-1) в уравнении (1) вычисляются из микроскопических сечений
отдельных изотопов и элементов (в барнах) по формуле:
где
ri
- ядерная плотность в смеси изотопа/элемента (барн-1см-1),
зависящая от координаты, T - температура,
которая также может меняться в пространстве. Аналогичная формула может быть
записана для сечения sX
. Это сечение включает процессы рассеяния и
деления, угловую зависимость которых обычно ограничивают только одной переменой
- косинусом угла рассеяния m0 = WЧWў.
Это позволяет разложить sX по полиномам Лежандра:
|
|
|
|
Интегрируя
по азимутальному углу и применяя теорему сложения, получим:
|
|
где
Требуемые
функционалы запишем в виде:
|
|
где
sr
- сечение реакции для этого функционала.
|
|
Для
того, чтобы получить групповое приближение, проинтегрируем уравнения (4) и (6)
в групповом интервале g. Получим,
соответственно:
и
|
|
|
|
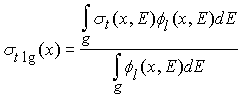 (10)
(10)
|
|
где
|
|
Последние три уравнения являются
основными определениями групповых сечения и матриц переходов. Отметим, что
групповые константы зависят как от сечений, данные о которых имеются в
библиотеках, так и от формы f
внутри группы.
2.2 Групповые
нейтронные константы
В
общей постановке задачи вид весовой функции f
не известен: ведь она по сути и является искомым распределением частиц. Однако
зачастую удается получить довольно точные значения констант, если в широких
энергетических интервалах с малым числом групп поток имеет характерные
зависимости (например, спектр деления, или 1/E, или спектр Максвелла). С другой
стороны, можно выбрать столь узкие группы, чтобы форма весовой функции не
оказывала существенного влияния на средние величины. Успех применения метода
групп во многом определяется умением находить баланс между выбором весовой
функции и числа групп.
Для задач переноса нейтронов задача
усложняется тем, что поток f(E)
обнаруживает глубокие провалы, вызванные поглощением в резонансах сечений. Эти
провалы могут существенно уменьшать (экранировать) скорость реакции s(E)f(E).
Для того, чтобы учесть этот эффект при получении констант, обычно используют
модель Бондаренко [9], особенно хорошо зарекомендовавшая себя в случае узких
резонансов и протяженных систем (для других случаев рекомендуется использовать
более строгий подход, основанный на решении детального уравнения замедления).
|
|
Согласно
этой модели, поток представляется в виде:
где
fl(E)- l-я
компонента разложения углового потока по полиномам Лежандра, Wl(E)- плавная функция
энергии (например, 1/E+спектр Уатта), st(E)
- полное макроскопическое сечение материала. Обычно зависимостью функции Wl(E)от l пренебрегают и оставляют за пользователем право выбора одной из
встроенных в программу функций C(E).
Предполагая, что суммарное влияние всех изотопов на самоэкранирование
выделенного i-изотопа можно
характеризовать одним параметром ![]() - "сечением разбавления", с учетом вышесказанного,
получим:
- "сечением разбавления", с учетом вышесказанного,
получим:
|
|
где
![]() - полное
микроскопическое сечение изотопа i.
- полное
микроскопическое сечение изотопа i.
Таким
образом, каждая компонента смеси имеет свою весовую функцию. Полное
макроскопическое сечение вычисляется по формуле:
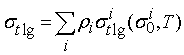 (15)
(15)
|
|
где
Аналогичные
выражения получаются для ![]() и
и ![]() .
.
|
|
Для
вычисления сечений разбавления в многокомпонентных средах обычно используется
итерационная процедура, построенная на уравнении:
где ri
- ядерная плотность или отношение концентраций.
|
|
В
области неразрешенных резонансов детальная зависимость сечений от энергий
неизвестна. Подынтегральные выражения заменяются в этом случае ожидаемыми
значениями:
где
ожидаемые значения являются результатом усреднения по распределениям ширин и
расстояний между уровнями в окрестности точки Е.
2.3 Источник деления
|
|
Сечение переноса sX
в уравнении (1) содержит все процессы, приводящие к возникновению нейтронов и
изменению их фазовых координат, включая реакции упругого и неупругого
рассеяния, (n,2n), деления и др. Иногда удобно отделить процесс деления от
остальных. Предполагая деление изотропным, источник деления в группе g запишем в виде:
где матрица межгрупповых переходов
определяется уравнением (12) с l=0.
Зависимость
спектра деления от начальной энергии нейтронов становится заметной лишь для
довольно высоких энергий; там, где ею можно пренебречь, справедливо выражение:
|
|
|
|
|
|
здесь
`ng
- выход нейтронов деления,sfg
- сечение деления, cg
- усредненный по начальной энергии нейтронов
спектр деления. Вновь определенные величины определяются формулами:
2.4 Константы
образования и переноса фотонов
Уравнение
переноса фотонов аналогично (7),если все входящие в него величины и определения
- потоки, сечения, группы отнести не к нейтронам, а к фотонам. Внешний источник
фотонов зависит от нейтронного потока и сечения образования фотонов в
нейтронных реакциях:
|
|
где
![]() определяется
уравнением (12), если заменить X на g.
определяется
уравнением (12), если заменить X на g.
Совместная задача переноса нейтронов и
фотонов может быть сведена к решению
уравнения (7), в котором фотонные группы рассматриваются как дополнительные
низкоэнергетические нейтронные группы. Если процессы (g,n)
не играют существенной роли, образованиe фотонов n®g
может рассматриваться аналогично процессу замедления нейтронов n®n.
Ниже схематически показана структура набора констант для решения обобщенного
уравнения переноса нейтронов и фотонов.
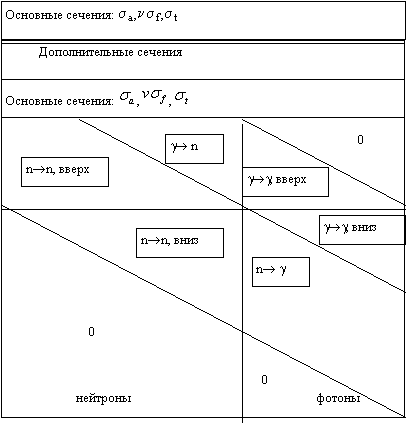
Рис.1
Структура набора констант.
3. БИБЛИОТЕКА
МИКРОКОНСТАНТ
Библиотека
групповых микроскопических данных GNPDL-30/19
включает следующие разделы:
-
данные о взаимодействии нейтронов с
веществом: сечения реакций, матрицы
межгрупповых переходов, - в области энергий 0.0001эВ - 20Мэв (30 групп);
-
матрицы образования фотонов в нейтронных
реакциях и данные о взаимодействии фотонов с веществом в области энергий от
0.01 - 20МэВ (19 групп);
-
кумулятивные и независимые выходы
осколков деления, усредненные по типичному спектру реактора на быстрых
нейтронах (1 группа).
3.1. Групповая
структура
Структура группового разбиения до
10.5МэВ практически совпадает с разбиением БНАБ, в верхней области добавлены
группы с тем, чтобы сохранить информацию, имеющуюся в файлах оцененных данных (
таблицы 1,2). В качестве весовой функции при усреднении групповых сечений
использовался стандартный спектр быстрого реактора
Таблица
1.
Структура нейтронных групп
N Границы групп Ширина Ширина
верхняя нижняя DE DU
МэВ
1 20.0000 - 17.0000 3.0000
1.1765
2 17.0000 - 15.0196 1.9804
1.1319
3 15.0196 - 13.9818 1.0378
1.0742
4 13.9818 - 10.5000 3.4818
1.3316
5 10.5000 - 6.5000 4.0000
1.6154
6 6.5000 - 4.0000 2.5000
1.6250
7 4.0000 - 2.5000 1.5000
1.6000
8 2.5000 - 1.4000 1.1000
1.7857
9 1.4000 - 0.8000 0.6000
1.7500
кэВ
10 800.0000 - 400.0000 400.0000
2.0000
11 400.0000 - 200.0000 200.0000
2.0000
12 200.0000 - 100.0000 100.0000
2.0000
13 100.0000 - 46.4159 53.5841
2.1544
14 46.4159 - 21.5443 24.8716
2.1544
15 21.5443 - 10.0000 11.5443
2.1544
16 10.0000 - 4.6416 5.3584
2.1544
17
4.6416 - 2.1544 2.4872
2.1544
18
2.1544 - 1.0000 1.1544
2.1544
эВ
19 1000.0000 - 464.1589 535.8411 2.1544
20 464.1589 - 215.4434 248.7155 2.1544
21 215.4434 - 100.0000 115.4434 2.1544
22 100.0000 - 46.4159 53.5841 2.1544
23 46.4159 - 21.5443 24.8715 2.1544
24 21.5443 - 10.0000 11.5443 2.1544
25 10.0000 - 4.6416 5.3584
2.1544
26 4.6416 - 2.1544 2.4872
2.1544
27 2.1544 - 1.0000 1.1544
2.1544
28 1.0000 - 0.4642 0.5358
2.1544
29 0.4642 - 0.2154 0.2487
2.1544
30 0.2154 - 0.0001 0.2153
-
Таблица
2.
Структура фотонных групп
N Границы групп Ширина Ширина
верхняя нижняя DE DU
МэВ
1 20.0000 - 17.0000 3.0000 1.1765
2 17.0000 - 15.0000 2.0000 1.1333
3 15.0000 - 13.0000 2.0000 1.1538
4 13.0000 - 11.0000 2.0000 1.1818
5 11.0000 - 9.0000 2.0000 1.2222
6
9.0000 - 7.0000 2.0000 1.2857
7
7.0000 - 5.5000 1.5000 1.2727
8
5.5000 - 4.5000 1.0000 1.2222
9
4.5000 - 3.5000 1.0000 1.2857
10
3.5000 - 2.5000 1.0000 1.4000
11 2.5000 - 1.7500 0.7500 1.4286
12 1.7500 - 1.2500 0.5000 1.4000
13 1.2500 - 0.7500 0.5000 1.6667
14 0.7500 - 0.3500 0.4000 2.1429
15 0.3500 - 0.1500 0.2000 2.3333
16 0.1500 - 0.0800 0.0700 1.8750
17 0.0800 - 0.0400 0.0400 2.0000
18 0.0400 - 0.0200 0.0200 2.0000
19 0.0200 - 0.0100 0.0100 2.0000
3.2. Типы
взаимодействий
Каждому
набору данных в архивах библиотеки GNPDL-30/19
поставлены в соответствие идентифицирующие номера: MAT - номер материала, MF -
номер типа данных, MT - номер типа взаимодействия. Номер материала MAT однозначно
определяет нуклид или элемент или вещество, к которому относятся эти данные. Он
в точности соответствует номеру,
принятому в библиотеке ENDF/B-6. Номер типа данных MF определяет структуру
записи данных с учетом типа налетающей частицы. Групповые данные могут быть векторного и матричного вида. Для их
хранения используются следующие структуры:
3 - групповые нейтронные сечения
6 - матрицы межгрупповых переходов для
нейтронов
16
- матрицы образования фотонов в нейтронных реакциях
23
- групповые сечения взаимодействия фотонов с веществом
26
- матрицы межгрупповых переходов для фотонов.
Внутри
каждого типа данные подразделяются по видам взаимодействия или другим характеристикам. Для идентификации этих
характеристик служит номер МТ.
Список
номеров материалов МAT и типов взаимодействий МТ, включенных в библиотеку GNPDL-30/19 приведен в Приложении А. Ниже приводится расшифровка значений МТ.
Суммы
процессов
1 (n,tot) 102 (n,g) 105 (n,t)
2 (n,n) 103 (n,p) 106 (n,He3)
4 (n,n') 104 (n,d) 107 (n,a)
Реакции
с возбуждением дискретных уровней ( N=0,1,2,3,...40 )
50
+N (n,n'N) 650+N (n,d'N) 750+N (n,He3'N)
600+N (n,p'N) 700+N (n,t'N) 800+N (n,a'N)
Реакции
с возбуждением уровней в области континуума
91 (n,n^) 699 (n,d^) 799 (n,He3^)
649 (n,p^) 749 (n,t^) 849 (n,a^)
Многочастичные
реакции
11 (n,2nd) 32 (n,nd) 108 (n,2a)
16 (n,2n) 33 (n,nt) 109 (n,3a)
17 (n,3n) 34 (n,nHe3) 111 (n,2p)
22 (n,na) 35 (n,nd2a) 112 (n,pa)
23 (n,n3a) 36 (n,nt2a) 113 (n,t2a)
24 (n,2na) 37 (n,4n) 114 (n,d2a)
25 (n,3na) 41 (n,2np) 115 (n,pd)
26 (n,np) 42 (n,3np) 116 (n,pt)
29 (n,n2a) 44 (n,n2p) 117 (n,da)
30 (n,2n2a) 45 (n,npa)
Процессы
деления
18 (n,ftot) 20 (n,nf)
19 (n,f) 21 (n,2nf)
22 (n,3nf)
Специальные
величины
452 полное число нейтронов деления
455 выход запаздывающих нейтронов
456 выход мгновенных нейтронов
Часть
данных, включенных в архив, отсутствует в библиотеке оцененных данных и
получена расчетным путем. Это данные следующих типов:
221
тепловое сечения рассеяния
251 средний косинус рассеяния (`m
)
252 средний логарифм потери энергии при
замедлении (`x
)
253 средний квадрат логарифмической потери
энергии (`g
)
301 керма-фактор, полученный из балансных
соотношений
443 керма-фактор, полученный из уравнений
кинематики (верхний предел)
444 сечение радиационного повреждения
Фотонные данные
содержат сечения и матрицы фото-атомных взаимодействий. В архив включены данные
для следующих элементов:
Имя МАТ Имя МАТ Имя МАТ Имя МАТ
01H 100 26Fe 2600 51Sb 5100 76Os 7600
02He 200 27Co 2700 52Te 5200 77Ir 7700
03Li 300 28Ni 2800 53I 5300 78Pt 7800
04Be 400 29Cu 2900 54Xe 5400 79Au 7900
05B 500 30Zn 3000 55Cs 5500 80Hg 8000
06C 600 31Ga 3100 56Ba 5600 81Tl 8100
07N 700 32Ge 3200 57La 5700 82Pb 8200
08O 800 33As 3300 58Ce 5800 83Bi 8300
09F 900 34Se 3400 59Pr 5900 84Po 8400
10Ne 1000 35B 3500 60Nd 6000 85At 8500
11Na 1100 36Kr 3600 61Pm 6100 86Rn 8600
12Mg 1200 37Rb 3700 62Sm 6200 87Fr 8700
13Al 1300 38Sr 3800 63Eu 6300 88Ra 8800
14Si 1400 39Y 3900 64Gd 6400 89Ac 8900
15P 1500 40Zr 4000 65Tb 6500 90Th 9000
16S 1600 41Nb 4100 66Dy 6600 91Pa 9100
17Cl 1700 42Mo 4200 67Ho 6700 92U 9200
18Ar 1800 43Tc 4300 68Er 6800 93Np 9300
19K 1900 44Ru 4400 69Tm 6900 94Pu 9400
20Ca 2000 45Rh 4500 70Yb 7000 95Am
9500
21Sc 2100 46Pd 4600 71Lu 7100 96Cm
9600
22Ti 2200 47Ag 4700 72Hf 7200 97Bk 9700
23V 2300 48Cd 4800 73Ta 7300 98Cf 9800
24Cr 2400 49In 4900 74W 7400 99Es 9900
25Mn 250 50Sn 5000 75Re 7500 100Fm 9920
Для
каждого элемента имеются данные:
501 полное взаимодействие
502 когерентное рассеяние
504 некогерентное рассеяние
516 образование пар
522 фотоэлектрическое поглощение
525 тепловыделение
Выходы осколков
деления приведены для следующих материалов:
90Th227 9025 92U0236 9231 94Pu241 9443 96Cm245 9640
90Th229 9031 92U0237 9234 94Pu242 9446 96Cm246 9643
90Th232 9040 92U0238 9237 95Am241 9543 96Cm248 9649
91Pa231 9131 93Np237 9346 95Am242M
9547 98Cf249 9852
92U0232 9219 93Np238 9349 95Am243 9549 98Cf251 9858
92U0233 9222 94Pu238 9434 96Cm242 9631 99Es254 9914
92U0234 9225 94Pu239 9437 96Cm243 9634 100Fm255
9936
92U0235 9228 94Pu240 9440 96Cm244 9637
Внутри
каждого материала они подразделяются на:
454 независимые выходы
459 кумулятивные выходы
Таблица
3. Кумулятивные выходы для важнейших материалов и
осколков деления
|
|
|
Th232 |
U233 |
U235 |
U238 |
Np237 |
Pu239 |
Pu240 |
Pu242 |
Am241 |
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
|
1 |
Kr-83 |
0.0211 |
0.0101 |
0.0053 |
0.0042 |
0.0034 |
0.0029 |
0.0025 |
0.0013 |
0.0017 |
|
2 |
Mo-95 |
0.0556 |
0.0634 |
0.0650 |
0.0511 |
0.0568 |
0.0481 |
0.0468 |
0.0378 |
0.0397 |
|
3 |
Tc-99 |
0.0283 |
0.0491 |
0.0610 |
0.0611 |
0.0661 |
0.0621 |
0.0596 |
0.0599 |
0.0662 |
|
4 |
Ru-101 |
0.0078 |
0.0317 |
0.0517 |
0.0614 |
0.0664 |
0.0601 |
0.0592 |
0.0609 |
0.0599 |
|
5 |
Ru-103 |
0.0024 |
0.0157 |
0.0303 |
0.0608 |
0.0577 |
0.0699 |
0.0596 |
0.0623 |
0.0622 |
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
|
6 |
Rh-103 |
0.0024 |
0.0157 |
0.0303 |
0.0608 |
0.0577 |
0.0699 |
0.0596 |
0.0623 |
0.0622 |
|
7 |
Rh-105 |
0.0017 |
0.0049 |
0.0096 |
0.0395 |
0.0264 |
0.0564 |
0.0565 |
0.0600 |
0.0664 |
|
8 |
Pd-105 |
0.0017 |
0.0049 |
0.0096 |
0.0395 |
0.0264 |
0.0564 |
0.0565 |
0.0600 |
0.0664 |
|
9 |
Pd-108 |
0.0020 |
0.0007 |
0.0005 |
0.0067 |
0.0177 |
0.0216 |
0.0326 |
0.0445 |
0.0425 |
|
10 |
Ag-109 |
0.0022 |
0.0003 |
0.0003 |
0.0035 |
0.0110 |
0.0147 |
0.0214 |
0.0350 |
0.0249 |
|
11 |
Cd-113 |
0.0023 |
0.0001 |
0.0001 |
0.0014 |
0.0003 |
0.0008 |
0.0010 |
0.0025 |
0.0019 |
|
12 |
In-115 |
0.0021 |
0.0001 |
0.0001 |
0.0012 |
0.0001 |
0.0004 |
0.0006 |
0.0011 |
0.0004 |
|
13 |
I-127 |
0.0021 |
0.0055 |
0.0015 |
0.0029 |
0.0018 |
0.0050 |
0.0035 |
0.0020 |
0.0058 |
|
14 |
I-135 |
0.0547 |
0.0503 |
0.0628 |
0.0677 |
0.0690 |
0.0654 |
0.0673 |
0.0738 |
0.0516 |
|
15 |
Xe-131 |
0.0176 |
0.0360 |
0.0289 |
0.0337 |
0.0315 |
0.0385 |
0.0334 |
0.0261 |
0.0356 |
|
16 |
Xe-135 |
0.0549 |
0.0625 |
0.0653 |
0.0683 |
0.0767 |
0.0760 |
0.0723 |
0.0749 |
0.0699 |
|
17 |
Cs-133 |
0.0411 |
0.0595 |
0.0669 |
0.0667 |
0.0647 |
0.0701 |
0.0671 |
0.0585 |
0.0550 |
|
18 |
Cs-134 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
|
19 |
Cs-135 |
0.0549 |
0.0626 |
0.0653 |
0.0683 |
0.0767 |
0.0762 |
0.0723 |
0.0749 |
0.0701 |
|
20 |
Nd-143 |
0.0646 |
0.0596 |
0.0595 |
0.0453 |
0.0503 |
0.0441 |
0.0448 |
0.0454 |
0.0367 |
|
21 |
Nd-145 |
0.0502 |
0.0344 |
0.0393 |
0.0371 |
0.0360 |
0.0298 |
0.0307 |
0.0343 |
0.0331 |
|
22 |
Pm-147 |
0.0569 |
0.0347 |
0.0449 |
0.0506 |
0.0500 |
0.0400 |
0.0424 |
0.0477 |
0.0406 |
|
23 |
Pm-148 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
|
24 |
Pm-149 |
0.0105 |
0.0077 |
0.0108 |
0.0160 |
0.0154 |
0.0121 |
0.0139 |
0.0159 |
0.0143 |
|
25 |
Sm-147 |
0.0284 |
0.0173 |
0.0224 |
0.0253 |
0.0250 |
0.0200 |
0.0212 |
0.0238 |
0.0203 |
|
26 |
Sm-149 |
0.0105 |
0.0077 |
0.0108 |
0.0160 |
0.0154 |
0.0121 |
0.0139 |
0.0159 |
0.0143 |
|
27 |
Sm-150 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
|
28 |
Sm-151 |
0.0034 |
0.0031 |
0.0041 |
0.0080 |
0.0074 |
0.0073 |
0.0085 |
0.0102 |
0.0083 |
|
29 |
Sm-152 |
0.0008 |
0.0021 |
0.0026 |
0.0053 |
0.0036 |
0.0057 |
0.0065 |
0.0080 |
0.0072 |
|
30 |
Eu-153 |
0.0003 |
0.0010 |
0.0015 |
0.0041 |
0.0019 |
0.0036 |
0.0045 |
0.0062 |
0.0060 |
|
31 |
Eu-154 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
|
32 |
Eu-155 |
0.0000 |
0.0002 |
0.0003 |
0.0014 |
0.0007 |
0.0016 |
0.0025 |
0.0031 |
0.0030 |
|
33 |
Gd-157 |
0.0000 |
0.0000 |
0.0000 |
0.0004 |
0.0002 |
0.0007 |
0.0009 |
0.0016 |
0.0016 |
3.3. Структура
микроконстант
Файл
MATXS предназначен для хранения сечений в виде векторов и матриц для всех
частиц, материалов и реакций; спектров запаздывающих нейтронов по временным
группам; константы тепловыделения и спектры фотонов при распаде.
Формат
MATXS хранит данные в компактном, удобном для доступа виде. Рабочая форма файла
- бинарная. Текстовые записи используются только для переноса данных. Описание
формата приведено в Приложении B.
Библиотека
микроконстант состоит из файлов. Имя файла однозначно определяет материал.
Длина его ограничена 6-ю символами. Обычно для естественной смеси изотопов имя
состоит из названия химического элемента (Fe,Cr,Ni); для отдельных изотопов к
названию химического элемента добавляется массовое число (Fe56,U235,Pu239); в
случае изомерии указывается изомерное состояние (Am242m).
Внутри
каждого файла данные распределены по типам частиц налетающих частиц. В
библиотеке MATXS-30/19 это нейтроны N и
фотоны G. В результате нейтронных реакций могут образовываться:
N
- нейтрон D - дейтрон A
- альфа (ядро 4He)
G
- фотон T - тритон B - бета частица
P
- протон H - гелион
(ядро 3He) R - остаточное
ядро
Материал
содержит данные различных типов, данные каждого типа могут дифференцироваться
по типам взаимодействия.
Имена
типов данных:
NSCAT рассеяние нейтронов
NG образование фотонов в нейтронных
реакциях
GSCAT рассеяние фотонов
NTHERM тепловые данные
PN образование нейтронов в протонных
реакциях
DKN данные по запаздывающим нейтронам
DKHG образование тепла при распаде и взаимодействия
фотонов
DKB данные по бета-распаду
Для
типов взаимодействия система образования имен довольно очевидна. Дадим здесь
расшифровку некоторых из них.
Символы
NWTO, NWT1, NTOT0,NTOT1 относятся к усредненным с весом потока и тока весовым
функциям и полным сечениям взаимодействия нейтронов. Данные по делению могут
быть дифференцированы по парциальным процессам: NF,NNF,N2NF; суммарное сечение
обозначается NFTOT. Сечения неупругого рассеяния на дискретных уровнях содержат
номер уровня: N01 - неупругое рассеяние на первом уровне; неупругое рассеяние
на континууме - NCN; cуммарное неупругое рассеяние - INELAS. Если уровень
распадается с излучением частиц, после номера уровня указываются имена частиц,
например: N22A, N28P,N29DAA.
Нейтронные
данные могут также содержать специальные величины.
HEAT
и DAME - сечения локального тепловыделения и энергии радиационного повреждения;
NUBAR,XI,GAMMA - параметры модели непрерывного замедления m,
x,
g;
INVEL - среднегрупповая обратная скорость; NU,NUD - среднее число нейтронов на
деление для мгновенных и запаздывающих, NUTOT - полное число нейтронов на
деление, аналогично CHI и СHID - для спектров нейтронов деления.
Для
фотонных взаимодействий используются имена:
GWTO - весовая функция, усредненная по
потоку,
GTOT0
- полное сечение взаимодействия
фотонов, нулевой момент
GCOH - когерентное рассеяние фотонов
GINCH - некогерентное рассеяние фотонов
GPAIR - образование пар
GABS - полное поглощение
GHEAT - тепловыделение.
3.4. Программы
сопровождения и доступа
Программа BBC [7] предназначена для выполнения различных
преобразований файлов. Ее возможности включают: изменение типа представления
(из бинарного - в текстовый и наоборот), объединение несколько материалов в
один файл, извлечения нужного материала из составного файла, получение
листингов и каталогов данных.
При
выполнении опции изменения типа представления, BBC читает
бинарный "matxs" или текстовый
"text", преобразует каждую запись из бинарного вида в текстовый или
из текстового в бинарный, и записывает новый текстовый "text" или
бинарный "matxs" файл.
Если
требуется получить листинг, BBC выдает записи на системное устройство вывода в
виде, удобном для чтения. Обычно
листинг выводится для выходного файла, если он не задан, выводится входной
файл. Признак "ird" указывает, какой из MATXS-файлов - входной или
выходной выдать в виде листинга.
Если
задано получение каталога, BBC формирует файл "index",
содержащий
в компактном виде листинг всех материалов, субматериалов и реакций, имеющихся в
выходном файле. Если выходной файл не задан, выдается каталог входного файла.
С помощью BBC можно модифицировать
бинарный MATXS файл, извлекая из него материалы или вставляя новые материалы из
бинарного файла "modinp". Новый файл при этом получает имя
"modout". Таким образом можно менять порядок материалов в файле.
Файл
из нескольких материалов может быть распакован на отдельные файлы, состоящие из
одного материала с изменением способа представления, если необходимо. Можно
изменить способ представления сразу целого набора файлов отдельных материалов
или объединить их в один файл, одновременно изменив тип представления и получив
листинг и каталог выходного файла.
Таким образом, программа BBC дает
возможность формировать компактные файлы и осуществлять их перенос из одной
системы программирования в другую.
Список используемых файлов:
matxs входной или выходной бинарный matxs
файл
text входной или выходной текстовый
matxs файл
modinp входной бинарный matxs файл для
модификации
modout выходной
бинарный или текстовый модифицированный matxs файл
index каталог библиотеки
Исходные
данные
карта 1
iinpf
тип входного файла
2
бинарный файл (с именем mat)
1
бинарный файл (matxs)
-1
текстовый файл (text)
-2
текстовый файл (с именем mat)
ioutf
тип выходного файла
2
бинарный файл (с именем mat)
1
бинарный файл (matxs или modout)
0
файл отсутствует
-1
текстовый файл (text)
-2
текстовый файл (с именем mat)
imodf
признак модификации
0
модификации отсутствуют (по умолчанию)
1
требуются модификации (modinp)
list
признак выдачи листинга
0
листинг не нужен (по умолчанию)
1
требуется листинг
indx
признак выдачи каталога
0
каталолг не нужен
1
требуется каталог (по умолчанию)
карта 2
(iinpf=2 или iinpf=-2)
diri
имя директории с входными файлами.
указать *name/*/ (не более 24
символов).
карта 3
(ioutf=2 или ioutf=-2)
diro
имя директории для выходных файлов.
указать *name/*/ (не более 24
символов).
карта 4
(ilist№0)
ird
признаки типа печати записи.
1
идентификатор файла
2
параметры файла
3
текстовое описание
4
данные файла
5
групповая структура
6
параметры материала
7
параметры вектора
8
блок векторов
9
параметры матрицы рассеяния
10
суб-блоки рассеяния
одно значение на каждый тип записи
0 - не печатать, 1 - печатать.
карта 5
(imodf№0)
hoper
тип модификации
s
выбрать материалы из входного matxs файла
i
ввести материалы из modinp файла
hmat1
первый материал в наборе
hmat2
последний материал (по умолчанию hmat2=hmat1)
повторить карту 3 пока не встретится пустая
строка
карта 6 (iinpf=2 или iinpf=-2)
hmati
список материалов в наборе, который нужно записать
в выходной файл matxs (в любом порядке)
карта 7 (только при модификации и дозаписи)
модификация применяется при imodf№0
при дозаписи ЅiinpfЅ=2
и ЅioutfЅ№2.
newver
номер версии нового файла
huser
строка-идентификатор нового пользователя
(не более 16 символов)
карта 8 (модификация или дозапись)
hline
новое текстовое описание данных (не более 72 символов)
повторить карту 8 для каждой строки нового
текстового блока данных.
Для
прекращения ввода ввести пустую строку. Если входные данные отсутствуют, старый
блок сохраняется прежним.
Программы доступа включают
в себя семейство процедур,
предназначенных для извлечения данных нужного типа из бинарного файла MATXS.
Эти функции, как правило, ориентированы на заполнение конкретной библиотеки
констант. Они учитывают особенность представления данных в этой библиотеке и
готовят для нее информацию соответствующим образом. Например, для заполнения
библиотеки групповых констант GNDL [8]
служат следующие процедуры.
ReadIsot:-
поиск данных для указанного нуклида и
запись требуемых таблиц в буферный массив с предварительной переработкой,
учитывающей структуру данных в библиотеке GNDL;
контрольные параметры данных заносятся
в область данных
COMMON
/BGETMATXS/ NG,NL,NT,NZ,EG(100),TEM(10),SIGZ(30)
где:
NG - число групп,
NL - максимальный
порядок представления индикатрисы рассеяния,
NT - число
температур,
NZ - число сечений
разбавления,
EG(NG+1) - массив
групповых границ (эВ)
TEM(NT) - массив температур (K),
SIGZ(NZ) - массив сечений разбавления (барн)
Буферный массив
определяется в вызывающей программе:
PARAMETER
(LBUF=1000000)
DIMENSION
BUF(LBUF),MBUF(LBUF)
EQUIVALENCE
BUF(1),MBUF(1)
и передается
функциям доступа месте с указанием максимальной длины LBUF.
Вызов процедуры:
call ReadIsot (HFILE,BUF,MBUF,LBUF)
где HFILE - имя
файла MATXS с данными для материала (переменная типа CHARACTER,
определенная в вызывающей программе)
GroupXS: извлечение таблицы сечений для заданной
группы IG. Таблица сечений - вектор XSR(16), позиции которого определены
следующим образом:
XSR(1)=NU - число нейтронов деления в группе IG
XSR(2)=SIGT - полное сечение
XSR(3)=SIGE - сечение упругого рассеяния
XSR(4)=SIGC - сечение
радиационного захвата
XSR(5)=SIGF - сечение деления
XSR(6)=SIGFNU - произведение
NU*SIGF
XSR(7)=SIGIN - сечение неупругого
рассеяния
XSR(8)=SGINZ - сечение неупругого
замедления
XSR(9)=SIGIN1 - 1-й момент сечения
неупругого рассеяния
XSR(10)=SGINZ1 - 1-й момент сечения неупругого замедления
XSR(11)=SIGN2N -
сечение реакции (n,2n)
XSR(12)=MU - средний косинус упругого рассеяния
XSR(13)=SGELZ - сечение упругого замедления
XSR(14)=XI - среднее значение изменения
летаргии в результате
упругого рассеяния
XSR(15)=SIGPOT -
сечение потенциального рассеяния ( не определено, =0) XSR(16)=CHI - спектр деления
Вызов процедуры:
call GroupXS(IG,XSR,BUF)
GroupFact: извлечение факторов
резонансной самоэкранировки для заданного
типа взаимодействия
HTYPE и группы IG - таблица XFACT (NZ,NT)
Тип взаимодействия - текстовая
переменная, определяющая тип
сечения, для которого требуются факторы
резонансной
самоэкранировки, имеющая допустимые значения:
TOT0 - полное сечение, усреднение по
потоку
TOT1 - полное сечение, усреднение по
току
ELAS - сечение упругого рассеяния
FISS
- сечение деления
CAPT - сечение радиационного захвата
INEL - сечение неупругого рассеяния
Зависимости от
температуры и сечения разбавления задаются в порядке возрастания значений
аргумента.
Вызов процедуры:
call GroupFact(IG,HTYPE,XFACT,BUF)
GroupMel: извлечение вектора переходов из группы IG в результате упругого
рассеяния - таблица XMATR(NG,NL)
Вызов процедуры:
call GroupMel(IG,XMATR,BUF)
GroupMin: извлечение вектора переходов из группы IG в результате неупругого рассеяния -
таблица XMATR(NG,NL)
Вызов процедуры:
call GroupMin(IG,XMATR,BUF)
ЛИТЕРАТУРА
1. P.F.Rose and
C.L.Dunford .”ENDF-102, Data Formats and Procedures for the Evaluated Nuclear
Data File, ENDF-6,” Brookhaven National
Laboratory report BNL-NCS-44945, (July 1990)
2. Г.Н.Мантуров,
М.Н.Николаев, А.М.Цибуля. "Система групповых констант БНАБ-93", ВАНТ,
серия "Ядерные константы", вып.1 (1996), с.59
3. R.E.MacFarlane et
al. “NJOY 94.10 Code System for
Producing Pointwise and Multigroup Neutron and Photon Sections form ENDF/B Data”. RSIC Peripheral
Shielding Routine Collection, PSR-355
4. D.E.Cullen et al. “The ENDF
Pre-Processing Code ( PRE-PRO 94)” Rep. IAEA-NDS-39, Rev.8,
Vienna (1994)
5. V.V.Sinitsa,A.A.Rineiskiy.
“GRUCON
- A Package of Applied Computer
Programs”, Rep. INDC(CCCP)-344, IAEA, Vienna(1993)
6. R.E.MacFarlane,
D.C.George. “UPD: A portable Version-Control
Program” Rep.LA-12057-MS
UC-705
7. R.E.MacFarlane
"TRANSX-2: A Code for Interfacing MATXS
Cross-Section Libraries to
Nuclear Transport Codes", LA-12312-MS (1992)
8.
A.V. Voronkov, V.I. Zhuravlev et al. “GNDL – a
Program System of Group Constants to Provide Calculations of Neutron and Photon
Fields”. Advances in Mathematics, Computations and Reactor Physics, Pittsburgh,
PA, USA, 1991
Приложение А.
Каталог библиотеки нейтронных данных
Имя МАТ Тип взаимодействия МТ
1H1 125 1,2,102
1H2 128 1,2,16,102
1H3 131 1,2,16
2He3 225 1,2,102-104
2He4 228 1,2
3Li6 325 1,2,4,24,51-81,102,103,105
3Li7 328 1,2,4,16,24,25,51-82,102,104
4Be9 425 1,2,16,102-105,107,600,650,700,701,800
5B10 525 1,2,4,51-85,102-104,107,113,600-605,800,801
5B11 528 1,2,4,16,22,28,51-60,91,102,103,105,107
6C 600 1,2,4,28,51-62,91,102-104,107,203,204,207
7N14 725 1,2,4,16,51-82,102-105,107,108,600-606,650-653,700,701,800-810
7N15 728 1,2,4,16,22,28,51-57,91,102-105,107
9F19 925 1,2,4,16,22,28,51-71,91,102-105,107
11Na23 1125 1,2,4,16,51-68,91,102,103,107
12Mg 1200 1,2,4,16,22,28,51-91,102,103,107
13Al27 1325 1,2,4,16,51-90,102-105,107,203,207
14Si 1400 1,2,4,16,22,28,51-72,91,102-104,107,203,207,600-614,649,
800-811,849
15P31 1525 1,2,4,16,28,91,102,103,107
16S 1600 1,2,4,16,22,28,51-91,102-105,107,111,203-205,207
16S32 1625 1,2,4,16,28,91,102,103,105,107
17Cl 1700 1,2,4,16,22,28,51-63,91,102,103,107
19K 1900 1,2,4,16,22,28,51-67,91,102,103,107
20Ca 2000 1,2,4,16,22,28,51-69,91,102-108,111,112
22Ti 2200 1,2,4,16,17,22,28,51-62,91,102-107,111,112,203,207
23V 2300 1,2,4,16,22,28,32,51-74,91,102-107,111,112
24Cr50 2425 1,2,4,16,22,28,51-56,91,102-104,107
24Cr52 2431 1,2,4,16,22,28,51-60,91,102,103,107
24Cr53 2434 1,2,4,16,22,28,51-63,91,102,103,107
24Cr54 2437 1,2,4,16,51-54,91,102,103,107
25Mn55 2525
1,2,4,16,22,28,51-79,91,102-107
26Fe54 2625 1,2,4,16,22,28,51-57,91,102-104,107
26Fe56 2631 1,2,4,16,22,28,51-75,91,102-107
26Fe57 2634 1,2,4,16,22,28,51-55,91,102,103,107
26Fe58 2637 1,2,4,16,22,28,51,52,91,102,103,107
27Co59 2725
1,2,4,16,22,28,32,33,51-69,91,102-108,111,112
28Ni58 2825 1,2,4,16,22,28,51-58,91,102-104,107
28Ni60 2831 1,2,4,16,22,28,51-61,91,102-104,107
28Ni61 2834 1,2,4,16,28,51-58,91,102,103,107,111
28Ni62 2837 1,2,4,16,22,28,51-54,91,102-104,107
28Ni64 2843 1,2,4,16,22,28,51,52,91,102-104,107
29Cu63 2925 1,2,4,16,22,28,51-72,91,102-104,106,107
29Cu65 2931 1,2,4,16,22,28,51-63,91,102-107
31Ga 3100 1,2,4,16,91,102,103,107
32Ge73 3234 1,2,4,51-54,91,102
32Ge74 3237 1,2,4,51-55,91,102
32Ge76 3243 1,2,4,51,52,91,102
33As75 3325 1,2,4,51-63,91,102
34Se74 3425 1,2,4,91,102
34Se76 3431 1,2,4,51-54,91,102
34Se77 3434 1,2,4,51-60,91,102
34Se78 3437 1,2,4,51-59,91,102
34Se80 3443 1,2,4,51-58,91,102
34Se82 3449 1,2,4,51-53,91,102
35Br79 3525 1,2,4,51-61,91,102
35Br81 3531 1,2,4,51-54,91,102
36Kr78 3625 1,2,4,16,51-53,91,102-107
36Kr80 3631 1,2,4,16,51-56,91,102-107
36Kr82 3637 1,2,4,16,51-59,91,102-107
36Kr83 3640 1,2,4,16,17,51-56,91,102-107
36Kr84 3643 1,2,4,16,51-63,91,102-107
36Kr85 3646 1,2,4,51,91,102
36Kr86 3649 1,2,4,16,17,51-64,91,102-105
37Rb85 3725 1,2,4,16,22,28,51-55,91,102-107,203-207
37Rb86 3728 1,2,4,51,91,102
37Rb87 3731 1,2,4,16,22,28,51-60,91,102-107,203-207
38Sr84 3825 1,2,4,91,102
38Sr86 3831 1,2,4,51-57,91,102
38Sr87 3834 1,2,4,51-57,91,102
38Sr88 3837 1,2,4,51-55,91,102
38Sr89 3840 1,2,4,51-62,91,102
38Sr90 3843 1,2,4,51-54,91,102
39Y89 3925 1,2,4,16,22,28,51-62,91,102-107
39Y90 3928 1,2,4,51,52,91,102
39Y91 3931 1,2,4,51-58,91,102
40Zr 4000 1,2,4,16,51-69,91,102,103,107
40Zr90 4025 1,2,4,16,51-55,91,102,103,107
40Zr91 4028 1,2,4,16,51-61,91,102,103,107
40Zr92 4031 1,2,4,16,51-56,91,102,103,107
40Zr93 4034 1,2,4,51-60,91,102
40Zr94 4037 1,2,4,16,51-58,91,102,103,107
40Zr95 4040 1,2,4,51-54,91,102
40Zr96 4043 1,2,4,16,51-54,91,102,107
41Nb93 4125
1,2,4,16,17,22,28,32,33,51-73,91,102-105,107
41Nb94 4128
1,2,4,51-58,91,102
41Nb95 4131
1,2,4,51-55,91,102
42Mo 4200 1,2,4,16,17,91,102
42Mo94 4231
1,2,4,51-57,91,102
42Mo95 4234
1,2,4,51-69,91,102
42Mo96 4237
1,2,4,51-56,91,102
42Mo97 4240
1,2,4,51-69,91,102
42Mo99 4246
1,2,4,51-56,91,102
43Tc99 4325 1,2,4,16,51-61,91,102
44Ru96 4425 1,2,4,91,102
44Ru98 4431 1,2,4,91,102
44Ru99 4434 1,2,4,51-62,91,102
44Ru100 4437
1,2,4,51-57,91,102
44Ru101 4440
1,2,4,51-69,91,102
44Ru102 4443
1,2,4,51-69,91,102
44Ru103 4446
1,2,4,51-64,91,102
44Ru104 4449
1,2,4,51,52,91,102
44Ru105 4452
1,2,4,51-59,91,102
44Ru106 4455
1,2,4,51-54,91,102
45Rh103 4525
1,2,4,16,51-64,91,102
45Rh105 4531
1,2,4,51-53,91,102
46Pd102 4625
1,2,4,91,102
46Pd104 4631
1,2,4,51-58,91,102
46Pd105 4634
1,2,4,51-63,91,102
46Pd106 4637
1,2,4,51-61,91,102
46Pd107 4640
1,2,4,51-65,91,102
46Pd108 4643
1,2,4,51-60,91,102
46Pd110 4649
1,2,4,51-62,91,102
47Ag107 4725
1,2,4,16,17,22,28,51-63,91,102-107,203-207
47Ag109 4731
1,2,4,16,17,22,28,51-70,91,102-107,203-207
47Ag111 4737
1,2,4,51-60,91,102
48Cd 4800 1,2,4,16,51-54,91,102,103,107
48Cd106 4825
1,2,4,91,102
48Cd108 4831
1,2,4,51-62,91,102
48Cd110 4837 1,2,4,51-57,91,102
48Cd111 4840
1,2,4,51-59,91,102
48Cd112 4843
1,2,4,51-56,91,102
48Cd113 4846
1,2,4,16,51-53,91,102,103,107
48Cd114 4849
1,2,4,51-56,91,102
48Cd116 4855
1,2,4,51-54,91,102
49In 4900 1,2,4,16,17,22,28,32,33,51-78,91,102-105,107
49In113 4925
1,2,4,51-56,91,102
50Sn112 5025
1,2,4,91,102
50Sn115 5034
1,2,4,51-54,91,102
50Sn116 5037
1,2,4,51-67,91,102
50Sn117 5040
1,2,4,51-55,91,102
50Sn118 5043
1,2,4,51-64,91,102
50Sn119 5046
1,2,4,51-58,91,102
50Sn123 5058
1,2,4,51-59,91,102
50Sn125 5064
1,2,4,51-53,91,102
50Sn126 5067
1,2,4,51-60,91,102
51Sb121 5125
1,2,4,51-57,91,102
51Sb123 5131
1,2,4,51-56,91,102
51Sb124 5134
1,2,4,51-58,91,102
51Sb125 5137
1,2,4,51-69,91,102
51Sb126 5140
1,2,4,51-55,91,102
52Te120 5225
1,2,4,91,102
52Te122 5231
1,2,4,51-54,91,102
52Te123 5234
1,2,4,51-58,91,102
52Te124 5237
1,2,4,51-61,91,102
52Te125 5240
1,2,4,51-56,91,102
52Te126 5243
1,2,4,51-63,91,102
52Te127m 5247
1,2,4,91,102
52Te128 5249
1,2,4,51-56,91,102
52Te129m 5253
1,2,4,91,102
52Te130 5255
1,2,4,51-60,91,102
52Te132 5261
1,2,4,51-56,91,102
53I129 5331 1,2,4,51-59,91,102
53I130 5334 1,2,4,91,102
53I131 5337 1,2,4,51-54,91,102
53I135 5349 1,2,4,91,102
54Xe124 5425
1,2,4,16,17,51-54,91,102-107
54Xe126 5431
1,2,4,16,17,51-54,91,102-107
54Xe128 5437
1,2,4,16,17,51-53,91,102-107
54Xe129 5440
1,2,4,16,17,51-56,91,102-105,107
54Xe130 5443
1,2,4,16,17,51-56,91,102-105,107
54Xe131 5446
1,2,4,16,17,51-56,91,102-105,107
54Xe132 5449
1,2,4,16,17,51-54,91,102-105,107
54Xe133 5452
1,2,4,51,91,102
54Xe134 5455
1,2,4,16,17,51-53,91,102-105,107
54Xe135 5458
1,2,4,51,91,102
54Xe136 5461
1,2,4,16,17,51-53,91,102-105,107
55Cs133 5525
1,2,4,16,51-55,91,102,103,107
55Cs134 5528
1,2,4,51-55,91,102
55Cs135 5531
1,2,4,51-53,91,102
55Cs136 5534
1,2,4,91,102
55Cs137 5537
1,2,4,51-62,91,102
59Pr141 5925
1,2,4,16,17,22,28,51-60,91,102-107
59Pr142 5928
1,2,4,91,102
59Pr143 5931
1,2,4,51-56,91,102
60Nd142 6025
1,2,4,51-58,91,102
60Nd144 6031
1,2,4,51-58,91,102
60Nd145 6034
1,2,4,16,17,22,28,51-64,91,102-107
60Nd146 6037
1,2,4,16,17,22,28,51-53,91,102-107
60Nd147 6040
1,2,4,51-53,91,102
60Nd148 6043
1,2,4,16,17,22,28,51-53,91,102-107
60Nd150 6049
1,2,4,16,17,22,28,51-59,91,102-105,107
61Pm148 6152
1,2,4,91,102
61Pm148m 6153
1,2,4,91,102
61Pm149 6155
1,2,4,51-56,91,102
61Pm151 6161
1,2,4,51-56,91,102
62Sm144 6225
1,2,4,91,102
62Sm148 6237
1,2,4,51-66,91,102
62Sm150 6243
1,2,4,51-66,91,102
62Sm151 6246
1,2,4,16,17,22,28,51-61,91,102-107
62Sm152 6249
1,2,4,16,17,22,28,51-64,91,102-107
62Sm153 6252
1,2,4,51-62,91,102
63Eu155 6337
1,2,4,16,17,22,28,51-59,91,102-107
63Eu156 6340
1,2,4,51-55,91,102
63Eu157 6343
1,2,4,91,102
64Gd152 6425
1,2,4,16,22,28,51-64,91,102,103,107
64Gd154 6431
1,2,4,16,22,28,51-64,91,102,103,107
64Gd156 6437
1,2,4,16,22,28,51-64,91,102,103,107
64Gd158 6443
1,2,4,16,22,28,51-64,91,102,103,107
64Gd160 6449
1,2,4,16,22,28,51-58,91,102,103,107
65Tb159 6525
1,2,4,51-66,91,102
65Tb160 6528
1,2,4,91,102
66Dy160 6637
1,2,4,51-64,91,102
66Dy161 6640
1,2,4,51-57,91,102
66Dy162 6643
1,2,4,51-56,91,102
66Dy163 6646
1,2,4,51-56,91,102
67Ho165 6725
1,2,4,16,17,51-63,91,102
68Er166 6837
1,2,4,51-69,91,102
61Pm147 6149
1,2,4,16,17,22,28,51-55,91,102-107
62Sm147 6234
1,2,4,16,17,22,28,51-64,91,102-107
62Sm149 6240
1,2,4,16,17,51-60,91,102,103,107
63Eu151 6325
1,2,4,16,17,51-67,91,102-107
63Eu152 6328
1,2,4,16,17,22,28,51-55,91,102-107
63Eu153 6331
1,2,4,16,17,51-60,91,102-107
63Eu154 6334
1,2,4,16,17,22,28,51-55,91,102-107
64Gd155 6434
1,2,4,16,22,28,51-64,91,102,103,107
64Gd157 6440
1,2,4,16,22,28,51-64,91,102,103,107
66Dy164 6649
1,2,4,16,17,51-62,91,102,103,107
68Er167 6840
1,2,4,51-63,91,102
71Lu175 7125
1,2,4,16,17,51-58,91,102,103,107
71Lu176 7128
1,2,4,16,17,51-58,91,102,103,107
72Hf 7200 1,2,4,16,51-62,91,102,103
72Hf174 7225
1,2,4,16,51-53,91,102,103
72Hf176 7231
1,2,4,16,51-53,91,102,103
72Hf177 7234
1,2,4,16,51-60,91,102,103
72Hf178 7237
1,2,4,16,51-53,91,102,103
72Hf179 7240
1,2,4,16,51-54,91,102,103
72Hf180 7243
1,2,4,16,51-53,91,102,103
73Ta181 7328
1,2,4,16,17,51-60,91,102,103
73Ta182 7331
1,2,4,16,17,51-58,91,102,107
74W 7400 1,2,4,16,17,28,51-91,102,103,107
74W182 7431
1,2,4,16,17,28,51-69,91,102,103,107
74W183 7434
1,2,4,16,17,28,51-64,91,102,103,107
74W184 7437
1,2,4,16,17,28,51-68,91,102,103,107
74W186 7443
1,2,4,16,17,28,51-68,91,102,103,107
75Re185 7525
1,2,4,16,17,51-59,91,102
75Re187 7531
1,2,4,16,17,51-66,91,102
79Au197 7925
1,2,4,16,17,51-63,91,102,103,107
82Pb206 8231
1,2,4,16,17,51-89,91,102,103,105,107
82Pb207 8234
1,2,4,16,17,51-89,91,102,103,105,107
82Pb208 8237
1,2,4,16,17,51-61,91,102,103,105,107
83Bi209 8325
1,2,4,16,17,22,28,32,33,51-69,91,102-107
90Th230 9034
1,2,4,16-18,51-67,91,102,452
90Th232 9040
1,2,4,16-18,51-65,91,102,452,455,456
91Pa233 9137
1,2,4,16-18,51-55,91,102,452
92U233 9222
1,2,4,16-18,51-54,91,102,452,455,456
92U234 9225
1,2,4,16-21,51-56,91,102,452,455,456
92U235 9228
1,2,4,16-21,37,38,51-84,91,102,452,455,456
92U236 9231
1,2,4,16-21,51-77,91,102,452,455,456
92U237 9234
1,2,4,16-18,91,102,452,455,456
92U238 9237
1,2,4,16-21,37,38,51-77,91,102,452,455,456
92U238 9237
1,2,4,16-21,37,38,51-77,91,102,452,455,456
93Np239 9352
1,2,4,16-18,51-58,91,102,452
94Pu236 9428
1,2,4,16-20,51-54,91,102,452
94Pu237 9431
1,2,4,16-20,51-60,91,102,452
94Pu238 9434
1,2,4,16-20,51-65,91,102,452,455,456
94Pu239 9437
1,2,4,16-21,37,38,51-81,91,102,452,455,456
94Pu240 9440
1,2,4,16-21,51-69,91,102,452,455,456
94Pu241 9443
1,2,4,16-18,51-65,91,102,452,455,456
94Pu242 9446
1,2,4,16-18,51-69,91,102,452
94Pu243 9449
1,2,4,16-18,37,91,102,452
94Pu244 9452
1,2,4,16-20,37,51-55,91,102,452
94Pu244 9452
1,2,4,16-20,37,51-55,91,102,452
95Am241 9543
1,2,4,16-18,51-68,91,102,452
95Am243 9549
1,2,4,16-20,37,51-67,91,102,452,455,456
96Cm241 9628
1,2,4,16-20,51-54,91,102,452
96Cm242 9631
1,2,4,16-20,51-53,91,102,452,455,456
96Cm244 9637
1,2,4,16-20,51-53,91,102,452
96Cm245 9640
1,2,4,16-18,37,91,102,452,455,456
96Cm246 9643
1,2,4,16-18,37,51-61,91,102,452
96Cm247 9646
1,2,4,16-18,37,91,102,452
96Cm248 9649
1,2,4,16-20,37,51-57,91,102,452
97Bk249 9752
1,2,4,16-18,51-68,91,102,103,107,452,455,456
98Cf249 9852
1,2,4,16-18,51-65,91,102,103,107,452,455,456
98Cf250 9855
1,2,4,16-18,37,91,102,452
98Cf251 9858
1,2,4,16-18,37,91,102,452,455,456
98Cf252 9861
1,2,4,16-18,37,91,102,452,455,456
Приложение B.
Формат MATXS
cs структура
файла
cs
cs тип записи условие наличия
cs ============================== ===============
cs заголовок файла всегда
cs параметры файла всегда
cs текстовое описание файла всегда
cs данные файла всегда
cs
cs
*************(в цикле по частицам)
cs
* групповая структура всегда
cs
*************
cs
cs
*************(в цикле по материалам)
cs
* параметры материала всегда
cs
*
cs
* ***********(в цикле по секциям материала)
cs
* * параметры векторов n1db.gt.0
cs
* *
cs
* * *********(в цикле по всем блокам векторов)
cs
* * * блок векторов n1db.gt.0
cs
* * *********
cs
* *
cs
* * *********(в цикле по всем блокам матриц)
cs
* * * параметры матриц n2d.gt.0
cs
* * *
cs
* * * *******(в цикле по всем блокам матриц)
cs
* * * * блок матриц n2d.gt.0
cs
* * * *******
cs
* * *
cs
* * * блок констант jconst.gt.0
cs
* * *
cs
*************
c
c--------------------------------------------------------------------
c
c
c--------------------------------------------------------------------
cr заголовок
файла
c
cl hname,(huse(i),i=1,2),ivers
c
cw 1+3*mult
c
cb format(4h 0v ,a8,1h*,2a8,1h*,i6)
c
cd hname
имя файла - matxs - (a8)
cd huse
имя пользователя (a8)
cd ivers
номер версии файла
cd mult
признак двойной точности
cd 1- a8 короткое слово
cd 2- a8 слово с двойной точностью
c
c--------------------------------------------------------------------
c
c
c--------------------------------------------------------------------
cr параметры
файла
c
cl npart,ntype,nholl,nmat,maxw,length
c
cw 6
c
cb format(6h 1d ,6i6)
c
cd npart
число частиц, для которых приведены групповые
cd данные
cd ntype
число типов данных
cd nholl
число слов в текстовом описании
cd nmat
число материалов
cd maxw
максимальная длина блока записи
cd length
длина файла
c
c--------------------------------------------------------------------
c
c
c--------------------------------------------------------------------
cr текстовое
описание
c
cl (hsetid(i),i=1,nholl)
c
cw nholl*mult
c
cb format(4h 2d /(9a8))
c
cd hsetid
текстовое описание набора (a8)
cd (до 72 позиций в строке)
c
c--------------------------------------------------------------------
c
c
c--------------------------------------------------------------------
cr данные
файла
c
cl
(hprt(j),j=1,npart),(htype(k),k=1,ntype),(hmatn(i),i=1,nmat),
cl
1(ngrp(j),j=1,npart),(jinp(k),k=1,ntype,(joutp(k),k=1,ntype),
cl
2(nsubm(i)i=1,nmat),(locm(i),i=1,nmat)
c
cw (npart+ntype+nmat)*mult+2*ntype+npart+2*nmat
c
cb format(4h 3d ,4x,8a8/(9a8)) hprt,htype,hmatn
cb format(12i6) ngrp,jinp,joutp,nsubm,locm
c
cd hprt(j)
текстовое имя частицы j
cd n
нейтрон
cd g
фотон
cd p
протон
cd d
дейтон
cd t
тритон
cd h
гелион ( ядро he-3 )
cd a
альфа ( ядро he-4 )
cd b
бета
cd r
остаточное ядро
cd (тяжелее чем альфа)
cd htype(k)
текстовое имя типа данных k
cd nscat
рассеяние нейтронов
cd ng
образование фотонов в нейтронных реакциях
cd gscat
рассеяние фотонов
cd pn
образование нейтронов в протонных реакциях
cd .
.
cd .
.
cd .
.
cd dkn
данные по запаздывающим нейтронам
cd dkhg
остаточное тепловыделение и образование фотонов
cd dkb
данные для бета-распада
cd hmatn(i)
текстовое имя материала i
cd ngrp(j)
число энергетических групп для частицы j
cd jinp(k)
тип налетающей частицы, связанный с данными типа k; для данных
типа dk jinp = 0.
cd joutp(k)
тип вылетающей частицы для данных типа k
cd nsubm(i)
число секций для материала i
cd locm(i)
адрес материала i
c
c--------------------------------------------------------------------
c
c
c--------------------------------------------------------------------
cr групповые
границы
c
cl (gpb(i),i=1,ngr),emin
c
cc ngr=ngrp(j)
c
cw ngrp(j)+1
c
cb format(4h 4d ,8x,1p,5e12.5/(6e12.5))
c
cd gpb(i)
верхняя граница энергии группы i для частицы j
cd emin
минимальная граница области энергий для частицы j
c
c--------------------------------------------------------------------
c
c
c--------------------------------------------------------------------
cr параметры
материала
c
cl
hmat,amass,(temp(i),sigz(i),itype(i),n1d(i),n2d(i),
cl
1locs(i),i=1,nsubm)
c
cw mult+1+6*nsubm
c
cb format(4h 5d ,a8,1p,2e12.5/(2e12.5,5i6))
c
cd hmat
текстовое имя материала
cd amass
атомный вес
cd temp
температура среды или другие параметры для
cd секции материала i
cd sigz
сечение разбавления или другие параметры для
cd секции материала i
cd itype
тип данных для секции материала i
cd n1d
число векторов для секции материала i
cd n2d
число блоков в матрице для секции материала i
cd locs
адрес секции материала i
c
c--------------------------------------------------------------------
c
c--------------------------------------------------------------------
cr параметры
векторов
c
cl
(hvps(i),i=1,n1d),(nfg(i),i=1,n1d),(nlg(i),i=1,n1d)
c
cw (mult+2)*n1d
c
cb format(4h 6d ,4x,8a8/(9a8)) hvps
cb format(12i6) iblk,nfg,nlg
c
cd hvps(i)
текстовые имена векторов
cd nelas упругое рассеяние нейтрона
cd n2n
(n,2n)
cd nnf
деление второго шанса
cd gabs
поглощение фотона
cd p2n
(p,2n)
cd .
.
cd .
.
cd .
.
cd nfg(i)
номер первой группы в векторе i
cd nlg(i)
номер последней группы в векторе i
c
c--------------------------------------------------------------------
c
c
c--------------------------------------------------------------------
cr блок
векторов
c
cl (vps(i),i=1,kmax)
c
cc kmax=суммарное число групп для векторов
блока j
c
cw kmax
c
cb format(4h 7d ,8x,1p,5e12.5/(6e12.5))
c
cd vps(i)
данные векторов блока j;
cd размер записи равен сумме числа
cd не превышающей maxw.
c
c--------------------------------------------------------------------
c
c
c--------------------------------------------------------------------
cr параметры
матриц рассеяния
c
cl hmtx,lord,jconst,
cl
1(jband(l),l=1,noutg(k)),(ijj(l),l=1,noutg(k))
c
cw mult+2+2*noutg(k)
c
cb format(4h 8d ,4x,a8/(12i6)) hmtx,lord,jconst,
cb jband,ijj
c
cd hmtx
текстовое имя блока
cd lord
число моментов
cd jconst
число групп с постоянным спектром
cd jband(l)
число групп в блоке l
cd ijj(l)
номер нижней группы в блоке l
c--------------------------------------------------------------------
c
c
c--------------------------------------------------------------------
cr блоки
матриц рассеяния
c
cl (scat(k),k=1,kmax)
c
cc kmax= сумма числа групп jband, принадлежащих
блоку,
cc умноженная на число моментов lord
c
cb format(4h 9d ,8x,1p,5e12.5/(6e12.5))
c
cw kmax
c
cd scat(k)
в записи элементы матриц объединяются по признаку
cd принадлежности к одной конечной
группе.
cd Порядок записи: переходы с моментом p0 в группу i,
cd переходы с моментом p1 в группу
i, ... , переходы
cd с моментом p0 в группу i+1, переходы с моментом p1
cd группу i+1, и т.д. Размер
записи равен сумме числа групп,
cd не превышающей maxw.
cd
cd Если jconst>0, вклады от jconst
нижних групп
cd приводятся отдельно.
c
c--------------------------------------------------------------------
c
c
c--------------------------------------------------------------------
cr блок
констант
c
cl
(spec(l),l=1,noutg(k)),(prod(l),l=l1,ning(k))
c
cc l1=ning(k)-jconst+1
c
cw noutg(k)+jconst
c
cb format(4h10d ,8x,1p,5e12.5/(6e12.5))
c
cd spec
нормированный спектр вылетающих частиц для исходной
cd частицы в группах с l1 по ning(k)
cd prod
сечения образования частиц (например, nu*sigf)
cd для начальных групп с l1 по ning(k)
cd
cd эта опция обычно используется для
независящих от энергии
cd спектров нейтронов и фотонов,
образующихся в процесах
cd деления и радиационного захвата, что
обычно имеет место
cd при низких энергиях.
c
c--------------------------------------------------------------------